Several tools are available that return stats and graphs from analysis of your fastq files and, inspired by a new paper just appeared on PLoS ONE, I just report want to report a couple of solutions that I found useful.
First is FastQC. This is a relative simple tools which take your fastq or bam/sam file and report all the essential stats you need to be sure that nothing has gone wrong with your sequencing. It's based on Java and so it can easily run on almost every platform without the need for tricky installation steps.
You can find this from the official web page at Babraham Bioinformatic Institute.
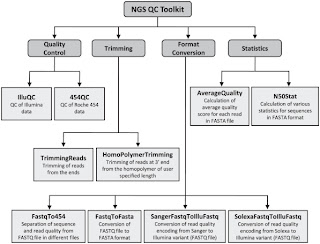
Second is NGS QC Toolkit. This is a set of tools for the quality control of next generation sequencing data. It accept data in the popular fastq format and provide with detailed results in the form of tables and graphs. Moreover it allows filtering of high-quality sequence data and includes few other tools, which are helpful in NGS data quality control and analysis (format conversion and trimming of the reads for example).
It is developed by the Indian National Institute of Plant Genome Research and you can find it at its official page here.
Also take a look at the official paper published on PLoS ONE in 2012 by Patel RK & Jain M
Third is this recent QC-chain tool that have cited above. The tool comprise a set of user-friendly tools for quality assessment and trimming of raw reads (Parallel-QC). Moreover it has an interesting feature that allows identification, quantification and filtration of unknown contamination to get high-quality clean reads. Authors stated that the tool was optimized based on parallel computation, promising that processing speed is significantly higher than other QC methods...This could be really useful if you routinely deal with a huge volume of data.
QC-chain is developed by the Computation Biology Team at Qingdao Institute of Bioenergy and Bioprocess Technology, and can be found here at the official web page.
This one also have an official paper published on PLoS ONE in 2013 by Zhou Q et al.


No comments:
Post a Comment